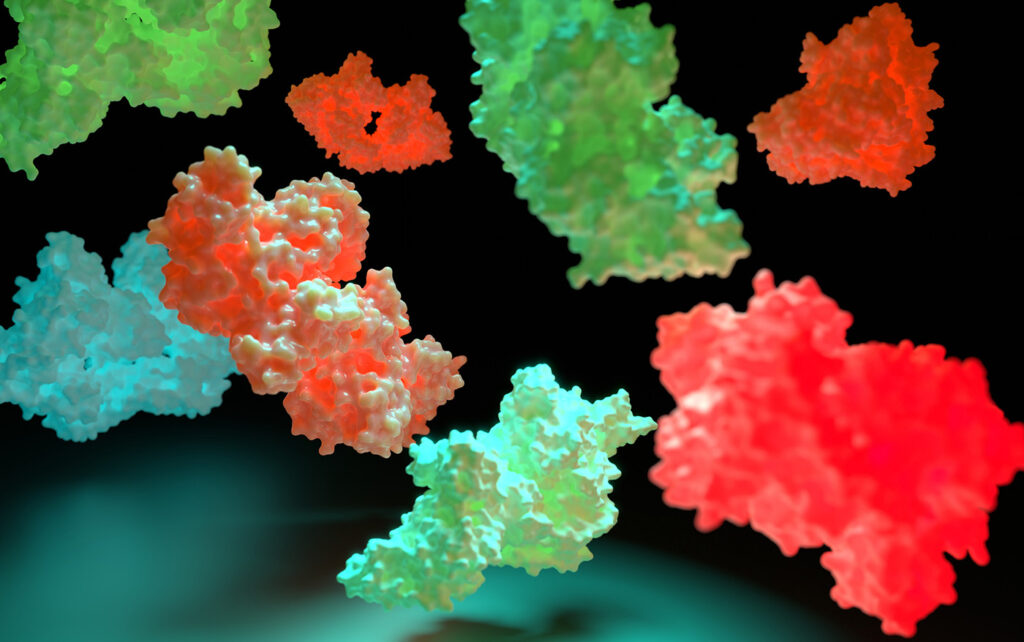
Credit: Miyako Nakamura / iStock / Getty Images Plus
The proteins found in nature represent only a tiny fraction of the nearly limitless protein structures that could theoretically exist. By designing proteins de novo—from scratch—scientists can more efficiently create novel therapeutics tailored for specific biological functions. In a new study, researchers introduce an AI-driven framework that accelerates this process, moving protein design toward custom therapeutics and precision medicine.
The study, from the European Molecular Biology Laboratory (EMBL), titled “AlphaDesign: a de novo protein design framework based on AlphaFold,” was recently published in Molecular Systems Biology.
“This paper is both a scientific validation of our approach and a foundation for our broader mission to transform and accelerate drug discovery and precision medicine by designing biology itself,” said Kashif Sadiq, PhD, founder and CEO of DenovAI.
Creating functional proteins
The German team began their study by generating raw sequences and structures of proteins to predict how the pieces change through binding. They tested their designed structures at multiple levels—from monomers, to dimers, to changes in conformation during binding—using a variety of established programs.
“Our method is versatile enough to design proteins with various properties, for example, proteins that are predicted to change conformation upon binding, as well as proteins predicted to bind multiple homologues of a protein, all without the requirement for model re-training or fine-tuning,” wrote the authors.
Testing revealed that the proteins designed by AlphaDesign “produce comparable or improved results to the state-of-the-art protein diffusion model, while representing a hallucination-based approach.”
“Accuracy is important because it reduces required design iteration, accelerating the path to clinical trials and enhancing the likelihood of improved efficacy, safety profiles, and clinical success,” said Sadiq.
But do they function in vivo?
To test whether their de novo protein designs could function in living systems, the team created active inhibitors targeting bacterial phage proteins. Typically, bacteriophage infection spurs the activation of RcaT toxins to inhibit bacterial growth, thus preventing infection. While the structure of one toxin, RcaT-Eco1, has been determined, the group aimed to explore the effectiveness of their strategy to create inhibitors for RcaT-Sen2, a variant with unconfirmed protein structure.
Treatment of E. coli colonies with inhibitor candidates showed that 19.3% of their designs significantly inhibited RcaT-Sen2 in vivo. The successful inhibitor design structures were confirmed using CD and NMR.
“AlphaDesign represents a new level of control in computational biology: the ability to generate new proteins with measurable, targeted function. This work demonstrates that AI can go beyond predicting biological structure to actively creating new molecular capabilities,” said Jan Korbel, PhD, head of data science and interim head of site at EMBL Heidelberg.
“We’ve demonstrated that AlphaDesign can generate proteins that fold as intended and function in living systems, including by targeting complex bacterial immune mechanisms,” said Sadiq.
This work represents a milestone in de novo protein creation, however, the researchers acknowledge that there is more work needed and more testing to be done. While the current work is a proof of concept for the utility of functional protein creation, there is much work to be done before it can be used for clinical applications.
“Ultimately, this translates to delivering better outcomes for patients,” concluded Sadiq. “Our vision is that for a given disease, we’ll one day be able to design the right therapeutic molecule, right from the start.”