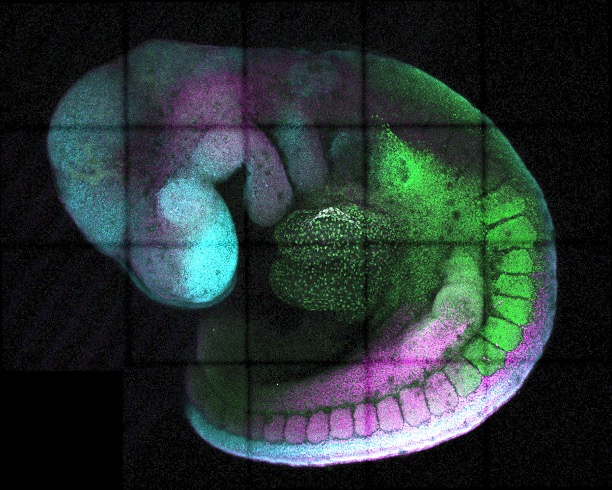
A developing mouse embryo expresses different genes (green, magenta, cyan) from head to tail, to generate different parts of the body [Irene Amblard, Development & Transcriptional Control Group, MRC Laboratory of Medical Sciences]
Scientists at the MRC Laboratory of Medical Sciences (LMS) have discovered a DNA-based “dimmer switch” that regulates the activity of a critical developmental gene, Cdx2. This work could pave the way for programmable gene expression strategies in therapeutics and disease modeling.
The findings of the study, published in Developmental Cell, “hint at the versatility of cis-regulatory elements, located throughout our genome, through precise changes to the DNA sequence,” Vicki Metzis, PhD, lead researcher, Development and Transcriptional Control group at LMS, told GEN. “Understanding this more generally could provide important advances for fine-tuning gene expression where and when it is needed for a range of applications.”
The paper titled “A dual enhancer-attenuator element ensures transient Cdx2 expression during posterior body formation” uncovers how the duration of expression of the gene Cdx2 is tightly regulated by a newly identified DNA sequence element called an “attenuator.”
“We have been particularly interested in Cdx2 for many years now, because of its critical role in embryonic development, including the formation of the spinal cord,” Metzis told GEN. “As Cdx2 plays such an important role, we wanted to understand what determines where and when a cell will express this critical factor.”
Cdx2 is expressed for a brief time during early development. The timing and duration of expression are pivotal to proper embryonic development. “Cdx2 is a key gene involved in shaping posterior body development, but its expression is extinguished really quickly,” first author Irène Amblard, PhD, told GEN. How the body controls this pulse of activity has remained unclear.
Using CRISPR-Cas9 gene editing, the team systematically dissected the regulatory landscape around the Cdx2 gene. They identified multiple cis-regulatory elements (CREs) that enhance Cdx2 expression in vitro. They further identified a DNA element within the Cdx2 locus that does not fit the classical definitions of either an enhancer or a silencer. Instead, this element modifies, or attenuates, expression with temporal and tissue specificity.
This attenuator modulates gene expression in a time- and cell-type-specific manner, unlike enhancers or silencers, which broadly switch genes on or off. Removing or mutating the attenuator significantly altered Cdx2 activity and disrupted normal body patterning in mouse embryos, leading to visibly malformed developmental structures.
The researchers found that they could reprogram this attenuator to act as an enhancer by modifying just a single motif within its sequence. “When we remove this region, we interfere with Cdx2 expression, and this impacts embryonic development,” Metzis explained to GEN. “What we also show is that by modifying the DNA sequence contained within the attenuator, we can reprogram the function of the element and its effect on Cdx2.”
To Amblard, one of the most striking moments came when comparing embryos with the attenuator knocked out. “Looking at the embryo generated using CRISPR-Cas9 knockout technology targeted against the attenuator and observing that the shape is different compared to wildtype embryos was a very striking result,” she shared with GEN. “It tells us that introducing subtle changes to the DNA can interfere with how an embryo develops.”
This dual behavior—able to enhance or attenuate gene expression depending on context—points to further complexity in the non-coding genome. “Our findings establish a dual cis-regulatory logic ensuring precise spatiotemporal control over gene expression for vertebrate body patterning,” the authors wrote.
The implications reach beyond developmental biology. By revealing how gene expression can be precisely tuned—not simply switched on or off—this work opens new possibilities for synthetic biology and gene therapy. Programmable control of gene activity could offer a new approach for treating diseases caused by gene misregulation, from developmental disorders to cancer.
“We’re excited because previous research suggests that our genome may harbor many different types of elements that finely tune gene expression, but they’ve not been easy to identify,” said Metzis. “If we can address this challenge, this holds enormous potential for unlocking new ways to treat diseases by fine-tuning gene expression where and when it’s needed.”
While more work is needed to explore the mechanisms through which this attenuator functions, Amblard said the next steps will focus on understanding how the element operates at a molecular level and what other genes may be similarly regulated.
As researchers move toward precision control of gene expression, discoveries like this provide key stepping stones. Not only do they deepen our understanding of how the genome builds a body, they also hint at how we might one day rewrite its instructions for therapeutic benefit.