In the intricate world of molecular biology, water has long been recognized as a fundamental player influencing the structure and function of proteins — the workhorse molecules of the cell. Despite its crucial role, the behavior and positioning of water molecules within protein environments have remained largely elusive to researchers, especially in the context of drug discovery. However, scientists at St. Jude Children’s Research Hospital have now unveiled a groundbreaking computational method, named ColdBrew, designed to illuminate the dynamic role of water molecules in protein binding sites. This innovative tool promises to dramatically refine our understanding of protein-ligand interactions and pave the way for more precise and efficient drug design.
Proteins are biological polymers composed of amino acids that fold into complex three-dimensional structures, critically influenced by their interaction with surrounding water molecules. These waters do not merely fill space; they participate actively in stabilizing the protein’s shape and modulating its biochemical activity. Particularly in drug discovery, where small molecules (ligands) are designed to bind specific protein sites to modulate function, knowing the exact location and behavior of water molecules is essential. Unfortunately, prevailing structural determination methods such as X-ray crystallography and cryo-electron microscopy operate at cryogenic temperatures, often distorting the natural positioning of water molecules due to freezing artifacts. This has led to an underappreciation and, in many cases, outright exclusion of water molecules in drug design efforts.
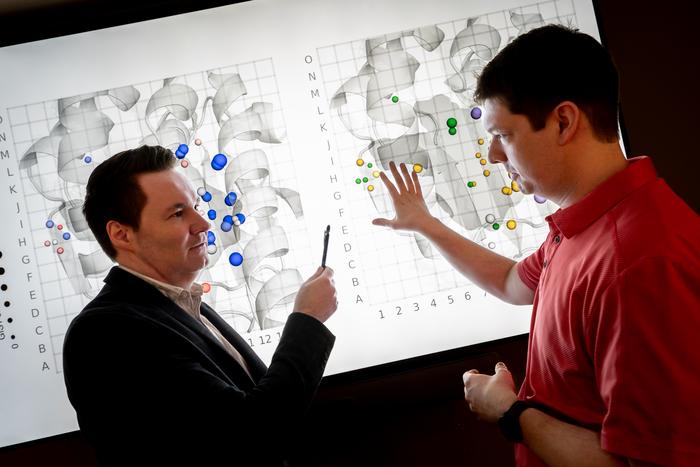
Recognizing this critical gap, Dr. Marcus Fischer and Dr. Justin Seffernick from St. Jude’s Department of Chemical Biology & Therapeutics developed ColdBrew, a computational algorithm that overcomes the limitations imposed by cryogenic structural data. Unlike conventional approaches, ColdBrew uses extensive protein water network data to calculate the likelihood of water molecule presence at physiological, higher temperatures. This correction allows researchers to better interpret experimental structures by distinguishing tightly bound, biologically relevant waters from those introduced artifactually by low-temperature data collection methods.
.adsslot_UJLv6ijRYZ{ width:728px !important; height:90px !important; }
@media (max-width:1199px) { .adsslot_UJLv6ijRYZ{ width:468px !important; height:60px !important; } }
@media (max-width:767px) { .adsslot_UJLv6ijRYZ{ width:320px !important; height:50px !important; } }
ADVERTISEMENT
The heart of ColdBrew lies in its ability to predict water displacement probabilities within protein structures, a feature with profound implications for drug discovery. Proteins bind ligands by displacing water molecules from their binding sites, but not all waters are equal; some are so tightly bound that displacing them is energetically unfavorable, while others readily vacate, facilitating ligand binding. By quantitatively assessing the likelihood that specific water molecules remain present at binding sites under native conditions, ColdBrew provides medicinal chemists with actionable insights. This enables the rational design of ligands that either exploit stable water molecules to enhance binding affinity or target sites where water displacement would be favorable, thus optimizing drug efficacy and selectivity.
One of the remarkable achievements of this project is the creation of a comprehensive, publicly accessible database containing ColdBrew predictions. Leveraging over 100,000 protein structures from the Protein Data Bank, the team conducted analyses covering more than 46 million water molecules. This expansive dataset offers an unparalleled resource for researchers around the globe, allowing them to tap into detailed water displacement predictions without the need for extensive computational resources. By democratizing access to these insights, ColdBrew has the potential to catalyze a paradigm shift in structure-based drug design, reducing trial-and-error in ligand development.
Beyond its immediate utility in pharmaceutical sciences, ColdBrew offers a methodological advancement with broad applicability across structural biology. The algorithm’s capacity to correct for cryo-induced artifacts elevates the fidelity of protein models, thereby enhancing downstream computational studies including molecular dynamics simulations and virtual screening. Importantly, the team demonstrated that the algorithm performs best at protein-ligand interfaces, the critical regions of interest for drug development, ensuring that its impact is maximally relevant to therapeutic innovation.
At the conceptual level, ColdBrew underscores the complex thermodynamic interplay between proteins, water, and ligands—a subtle dance that governs molecular recognition. Water molecules, often dismissed as inconvenient noise in structural data, emerge as critical determinants of biochemical specificity and affinity. The algorithm’s predictive capacity thus illuminates the “hidden” water landscape, allowing scientists to factor in water-mediated interactions hitherto considered too challenging to characterize reliably.
Moreover, ColdBrew encourages a reevaluation of prevailing drug discovery strategies that frequently disregard water molecules due to the uncertainty of their positioning. These findings suggest that drug designers may have unknowingly avoided targeting binding sites with tightly bound water molecules, potentially missing opportunities for improved binding or altered pharmacodynamics. Armed with ColdBrew’s insights, the design process becomes more nuanced, balancing displacement and accommodation of water molecules to fine-tune ligand efficacy.
From a technical standpoint, developing ColdBrew involved sophisticated analysis of temperature-dependent protein-water interactions. The algorithm probabilistically models water occupancy based on structural data obtained under varying temperature regimes, integrating these with known principles of water thermodynamics and protein chemistry. This methodological innovation bridges experimental and computational fields, harnessing large-scale structural data to resolve a long-standing bottleneck in capturing the true aqueous environment of proteins.
Collaboration with the broader scientific community is a key aspect of the ColdBrew initiative. Recognizing the importance of open science, the researchers have made their predictions and underlying datasets accessible via a digital repository, facilitating integration with existing bioinformatics pipelines. This openness accelerates validation efforts, adoption, and iterative improvement of the tool as more data becomes available.
The pioneering work on ColdBrew was supported by funding from the National Institutes of Health and the American Lebanese Syrian Associated Charities, reflecting the vital interplay between basic science and translational research. Dr. Fischer and his team at St. Jude Children’s Research Hospital continue to push the boundaries of chemical biology, employing cutting-edge computational methods to unravel complexities that have long challenged researchers in the realm of protein structure and function.
In conclusion, the introduction of ColdBrew represents a transformative step in structural biology and drug discovery, addressing a crucial blind spot by bringing water molecules into sharper focus. As drug developers seek increasingly sophisticated ways to modulate biological targets, tools like ColdBrew that reveal the nuanced behavior of water will undoubtedly become indispensable. By redefining how researchers interpret protein structures, ColdBrew not only enhances molecular insight but also promises to accelerate the discovery of safer and more effective therapeutics.
Subject of Research: The role of water molecule dynamics in protein structures and their implications for drug discovery.
Article Title: ColdBrew: A Novel Algorithm for Accurate Water Displacement Predictions in Protein Structures Enhancing Drug Design.
News Publication Date: June 27, 2025.
Web References:
ColdBrew Data Repository
Fischer Lab at St. Jude
Department of Chemical Biology & Therapeutics
St. Jude Children’s Research Hospital
Image Credits: St. Jude Children’s Research Hospital
Keywords
Drug discovery, Water molecules, Protein structure, Protein-ligand binding, Cryogenic temperature artifacts, Computational biology, Structural biology, Protein Data Bank, Molecular dynamics, Chemical biology, ColdBrew algorithm, Thermodynamics
Tags: biochemical activity modulationColdBrew computational methodcomputational drug design toolscryo-electron microscopy challengesdrug discovery innovationsmolecular biology breakthroughsprotein structure and functionprotein-ligand interactionsSt. Jude Children’s Research Hospitalstructural determination methodswater dynamics in proteinsX-ray crystallography limitations