Sponsored content brought to you by
When using antibodies in research or therapeutics, scientists focus on how these proteins bind to their intended targets, such as antigens or immunogens. That binding process depends on many factors. “One often overlooked parameter is the exact binding site of your antibody on the protein of interest, also known as the epitope,” says Danielle Callahan, director of global product management at Proteintech. “Carefully choosing your antibodies based on epitope binding site can greatly influence experimental outcomes, therapeutic strategies, and diagnostic accuracy.” Finding that specific binding site is called epitope mapping, and Proteintech unveiled a significant advance in this process.
Recently, Proteintech was the first to release epitope mapping data at scale on its website. Knowledge about an antibody’s epitope binding site is beneficial to researchers for many reasons, as recently described in a blog post by Callahan.
For example, post-translational modifications can impact epitope mapping. “If an epitope is near a phosphorylation, glycosylation, or acetylation site, modifications may alter binding, affecting experimental results,” Callahan explained. “Alternatively, you may want an antibody that binds selectively to a post-translational modification, such as a specific phosphorylation site.”
When using multiplex assays like flow cytometry or mass spectrometry, more than one epitope must be considered, because multiple antibodies are used to simultaneously detect different proteins. In these cases, “knowing the epitopes is vital for preventing interference,” Callahan noted. “By selecting antibodies that target non-overlapping epitopes, researchers can ensure that each antibody binds specifically to its target, allowing for clearer and more accurate data.”
The location of the epitope can also impact results, especially when developing an antibody-based therapy. “Epitope location affects whether an antibody blocks receptor-ligand interactions or activates/inhibits signaling pathways in therapeutic research,” Callahan pointed out. “Identifying these epitopes is imperative for developing an effective treatment.”
More to see in 3D
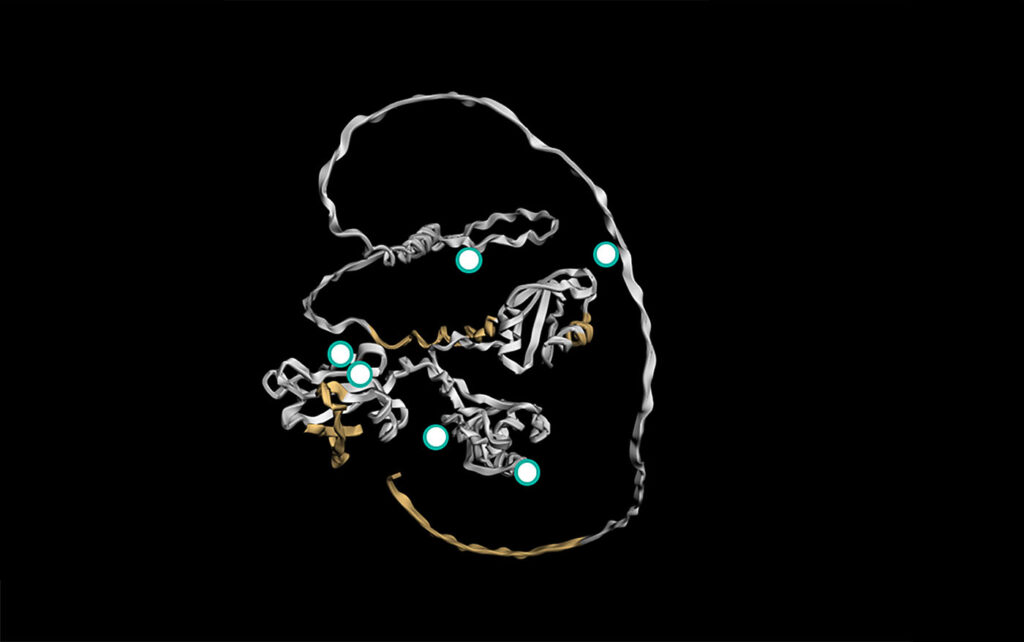
Like all biological structures, epitopes are three-dimensional. So, in March 2025, Proteintech announced its 3D Epitope Mapping. This pioneering project is part of an advanced validation initiative to produce 3D maps of the epitopes on target proteins by combining published protein data and modeling based on artificial intelligence (AI). With this technology, a scientist can quickly see a 3D representation of an antibody’s target, especially the epitope. As a result, the visual display allows for more precision in choosing an antibody for a specific application.

Proteintech is building a collection of epitopes that can be viewed in 3D. “We plan to have our entire catalog mapped in the near future,” says Jason Li, PhD, an immunologist and Proteintech’s CEO. “With more epitopes mapped, we hope the days when scientists have to try multiple antibodies for one experiment are behind us.”
In Proteintech’s expanding collection of 3D visualizations of targets for the company’s antibodies, the precise locations of the related epitopes can be explored. For example, the epitope is clearly marked by labeling the specific amino-acid residues at both ends.
Overall, Proteintech’s 3D Epitope Viewer improves efficiency in the development and application of antibodies, which will benefit academic and industrial scientists. “Whether in research, diagnostics, or therapeutics, knowing where an antibody binds on its target protein is indispensable for achieving precise, reproducible, and meaningful results,” Callahan concluded. “Therefore, characterizing the epitope is not just a technical step—it is essential for the success of any experiment that relies on antibodies.”
Find out more about 3D Epitope Mapping on ptglab.com.