Tapping into the structural intricacies of the cell nucleus was once a pipe dream, Jason Buenrostro, PhD, told me when reflecting on the early beginnings of the spatial genomics field. When he started his lab as an institute member at the Broad Institute eight years ago, sifting through millions of DNA sequences to interrogate new disease-relevant genes was arguably one of the “most important things happening in biology.”
Yet, the 2D sequences of As, Ts, Cs, and Gs were far from the highly structured and compact genome in its natural form, where spatial organization guided a stew of protein interactions and epigenetic marks needed for gene regulation and function.
This high-resolution vision has recently taken one step forward. Buenrostro, who is also an associate professor in the Department of Stem Cell and Regenerative Biology at Harvard University, has now developed a new technique that visualizes how structural disruptions in the nucleus can impact health and disease at nanoscale resolution.
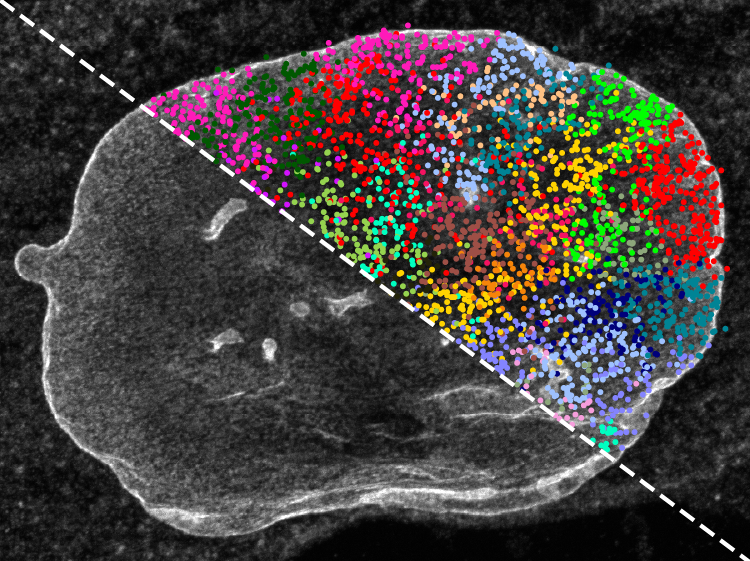
The imaging method, termed expansion in situ genome sequencing (ExIGS), enables both sequencing of genomic DNA and identification of super-resolution localization of nuclear proteins in a single cell. The work was published as a paper in Science titled, “Expansion in situ genome sequencing links nuclear abnormalities to aberrant chromatin regulation.”
“When we think of cell biology, we usually think of imaging and sequencing as two very different modalities,” said Zachary Chiang, PhD, a postdoctoral researcher in the Buenrostro group and co-first author of the study. “This technology is a way of connecting the types of images clinicians use to diagnose disease with high-resolution molecular readouts—and we hope it will allow scientists to ask new types of questions about disease.”
The team applied ExIGS to fibroblasts from patients with progeria, a rare and progressive genetic disorder marked by accelerated aging and caused by mutations in lamin proteins, known to provide crucial structural components to support and stabilize the nucleus. Results showed that progeria-derived fibroblasts displayed distorted nuclei with invaginations linked to hotspots of abnormal chromatin repression associated with disease.
Notably, the authors also observed invaginations in cells from a 92-year old individual without the condition, suggesting that these changes in lamin structure disrupt gene expression in both progeria and the aging process.
While gene repression in the periphery of the nucleus has been well studied, repression near the center of the nucleus, where invaginations form, is less understood. The study suggests that the spatial organization of the genome deep in the nucleus could be an underappreciated factor controlling gene expression throughout a person’s lifetime.
Buenrostro sees ExIGS as a tool that provides expanded access to fundamental biology and launching points for new applications. As an example, the spatial insights linking genome structure distortion and progeria can inspire new diagnostic tools focused on nuclear morphology.
Two people with crazy ideas
Buenrostro developed ExIGS in collaboration with Broad colleague and fellow associate professor at Harvard, Fei Chen, PhD. The spatial pioneers originally connected in 2015 as two people with “no experience who came up with some crazy ideas about how we might change biology,” remarked Buenrostro in an interview with GEN.
At the time, both Chen and Buenrostro were fresh PhD graduates who were awarded full support to establish their own independent research labs by the Schmidt Fellows Program at the Broad Institute. Chen had just developed a method, termed expansion microscopy, which expanded tissues using a polymer gel to allow typical light microscopes to capture high-resolution images of biological structures.
Five years later, Chen and Buenrostro, in collaboration with Ed Boyden, PhD, professor in the Department of Brain and Cognitive Sciences at Massachusetts Institute of Technology (MIT), developed in situ sequencing, which made the spatial advance of sequencing DNA within intact cells. While alternative methods had allowed scientists to reconstruct structural information about the genome, in situ sequencing was the first technology that gave scientists a direct look at the genome in its native environment.
ExIGS was born from a marriage of these two tools to push the spatial resolution and sequencing capabilities to nanoscale. While in situ sequencing was best suited for analyzing cells with large nuclei, such as embryos, ExIGS’s deepened resolution expands genome visualization capabilities to additional tissues and cell types to reach more disease-relevant contexts.
As next steps, the research team is looking at naturalistic aging with a particular focus on how therapeutics based on OSKM (Oct4, Sox2, Klf4, and Myc), a set of four transcription factors that are crucial for reprogramming cells into induced pluripotent stem cells, can rejuvenate nuclear structure. The group also hopes to expand ExIGS capabilities to RNA sequencing and chromatin accessibility.