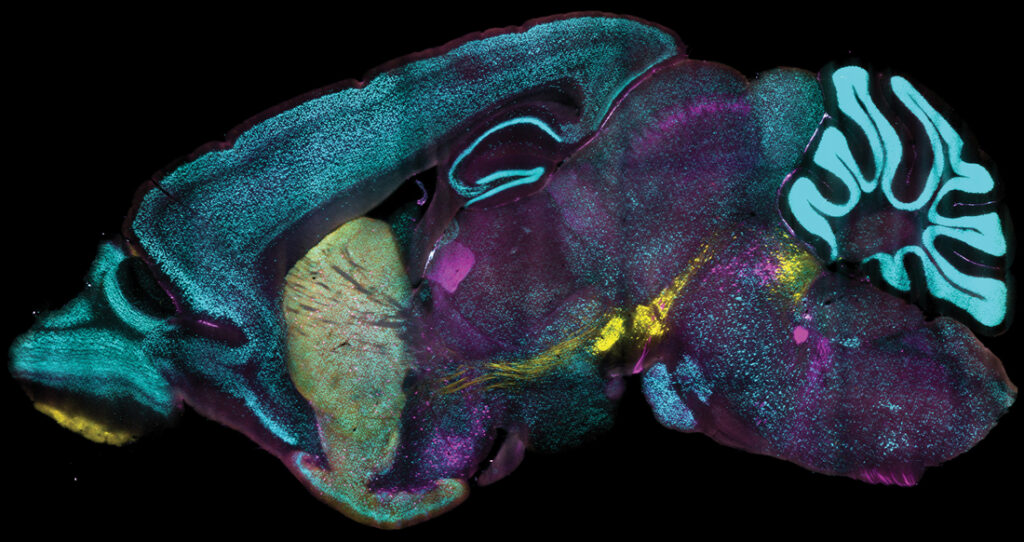
MIT researchers have unveiled new technology that allows for the labeling of proteins in millions of individual cells within fully intact tissues. The method, named Continuous Redispersion of Volumetric Equilibrium (CuRVE), enables this labeling process to occur quickly and uniformly, making it possible to label large tissue samples, like entire rodent brains, in a single day.
This research is detailed in a study published in Nature Biotechnology, entitled “Uniform volumetric single-cell processing for organ-scale molecular phenotyping.” The work demonstrates a transformative step forward in profiling protein expression across whole, dense tissues.
“Conventionally, investigating the molecules within cells requires dissociating tissue into single cells or slicing it into thin sections, as light and chemicals required for analysis cannot penetrate deep into tissues,” said senior author Kwanghun Chung, PhD, associate professor at the Picower Institute for Learning and Memory and MIT.
The Chung lab is no stranger to producing protein visualization breakthroughs. “Our lab developed technologies such as CLARITY and SHIELD, which enable investigation of whole organs by rendering them transparent,” Chung said.
Last year the team shared a new imaging technique that enables high-resolution, high-throughput imaging of human brain tissue without destroying the tissue using the mELAST sectioning and staining method and computational system UNSLICE to seamlessly stitch together the resulting 3D images.
“We now needed a way to chemically label whole organs to gain useful scientific insights,” Chung pointed out.
He explained the new method with an analogy: “Imagine marinating a thick steak by simply dipping it in sauce. The outer layers absorb the marinade quickly and intensely, while the inner layers remain largely untouched unless the meat is soaked for an extended period. The same principle applies to chemical processing of tissues: if cells within a tissue are not uniformly processed, they cannot be quantitatively compared.”
A prime challenge in labeling large tissue samples is the time it takes for antibodies, the so-called marinade, to penetrate the sample completely. “It can take weeks for these molecules to diffuse into intact organs, making uniform chemical processing of organ-scale tissues virtually impossible and extremely slow,” said Chung. The team developed the CuRVE framework to address this very issue.
CuRVE continuously maintains a dynamic equilibrium of the tissue’s chemical environment, allowing for uniform antibody dispersion and labeling across the entire tissue. The implementation of CuRVE in this study integrates innovations that control antibody binding speed and enhance their penetration into tissues. They developed a computational simulation, allowing them to test different parameters including changes in binding rates and tissue densities virtually.
The team at MIT built on top of prior successes to implement their approach in real samples. They began by using a previous method called SWITCH, in which antibodies are temporarily turned off, allowing them to penetrate into tissues before binding is reactivated. SWITCH has limited applications, that prevent ongoing treatment due to the harsh chemicals used in this method.
By screening a library of chemicals, the researchers identified deoxycholic acid as an ideal candidate for continuously modulating antibody binding speed. They also optimized the labeling process by varying the pH or acid concentration and employing another technique developed by the Chung lab called stochastic electrotransport, a method used to accelerate antibody dispersion using electric fields.
Through the marriage of the techniques of delaying antibody activation and modifying the speed of antibody dispersion through a sample using the CuRVE framework, eFLASH was born.
The team successfully labeled large tissue samples, including whole mouse and rat brains, whole mouse embryos, and human tissue blocks with eFLASH. They used over 60 different antibodies in their tests and each sample was also labeled within a day—when more widely used methods often take days or weeks. Additionally, the process required no re-optimization across different preparations, demonstrating its robustness and versatility.
“This is a significant leap, especially in terms of the actual performance of the technology,” commented co-lead author Dae Hee Yun, PhD, application engineer at LifeCanvas Technologies and former MIT graduate student.
In comparative tests, eFLASH’s labeling precision outperformed cells genetically engineered to produce fluorescence for a protein of interest. With genetic labeling, the fluorescent proteins are not always produced in sync with the target protein, and the fluorescent proteins may be expressed with a delay and persist longer in the cell than the target protein, reducing the accuracy of genetic labeling.
The team used eFLASH to label genetically engineered cells as a visual comparison of target protein expression. Using parvalbumin (PV) expressing cells, they “discovered highly variable regionalized reduction of PV immunoreactive cells in wild-type adult mice, a phenotype missed by the commonly used genetic labeling.” In short, each method shows both overlap and discrepancies in target protein expression in cells that are labeled using genetic engineering and eFLASH.
Yun noted that the two methods are “two different tools for the job.”
The authors emphasized the complementary nature of genetic and antibody-based labeling approaches, suggesting that integrating both methods may provide a more nuanced understanding of protein expression. As the authors wrote, “Our discovery of large regionalized loss of PV-immunoreactive neurons in healthy adult mice and with high individual variability emphasizes the importance of holistic and unbiased phenotyping.”
The CuRVE framework and its implementation in eFLASH mark a significant advancement in volumetric single-cell analysis. By enabling comprehensive protein labeling in large samples, this technology has the potential to facilitate more insights into the fields of neuroscience and beyond.