Recent discoveries have illuminated the intricate relationship between fungal viral infections and their viral hosts, particularly focusing on the less understood mechanisms in fungus-virus systems. The research conducted by a team from Japan, led by Associate Professor Shinji Honda of the University of Fukui, sheds light on how the fungus Neurospora crassa modulates its antiviral responses through RNA editing mechanisms. This intricate biological interplay captures the attention of both microbiologists and virologists as it has the potential to offer new insights into genetic engineering applications in mycology.
The importance of understanding the symptom expression in viral infections arises from its complex nature tied to the molecular dialogs that occur between the virus and its host. This dialogue is particularly elusive within fungal systems, where many mycoviruses can inhabit their hosts asymptomatically. Through extensive genetic studies, researchers have attempted to uncover the specific genes and pathways that lead to either symptom induction or suppression in these systems. The discovery of how neighboring genes can impact viral responses represents a significant advancement in our understanding of fungal virology.
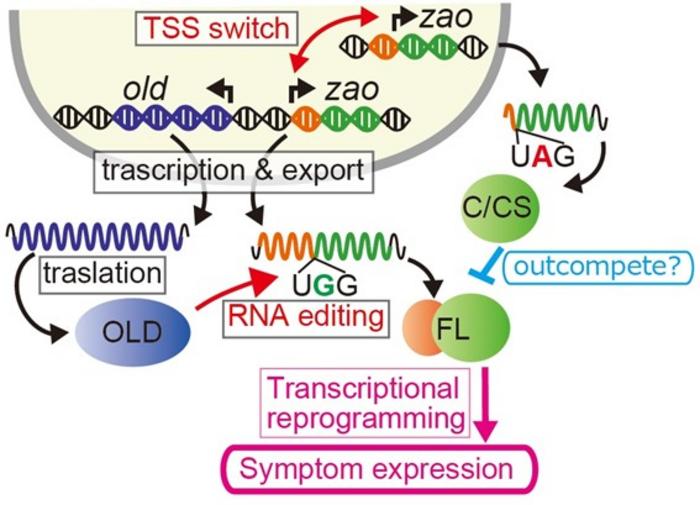
Honda’s research team utilized a novel fungal virology system unique to N. crassa to unravel the genetic mechanisms behind symptom induction when faced with viral infections. Their pivotal findings, published in the prestigious journal Cell Host & Microbe, highlight the ability of RNA-editing enzymes to alter the message carried by specific mRNAs of neighboring genes known to be integral in the host’s antiviral response. The compelling nature of their findings emerged from their previous isolations of viruses, including the asymptomatic Neurospora crassa fusarivirus 1 (NcFV1).
The RNA editing conducted by A-to-I enzymes on the fungal mRNA transcripts represents a critical mechanism where the editing modifies the expression of master transcription factors. These transcription factors play an essential role in orchestrating the fungal defense mechanisms against viral adversities, leading to enhanced antiviral responses. This remarkable strategy showcases a level of bioengineering within the fungal kingdom, illustrating how fungi adapt at a molecular level to the pressures exerted by viral infections.
When analyzing the genetic modifications, the researchers identified an interplay between the genes labeled old and zao. The gene old, which encodes for deaminase enzymes, appears to modify the transcripts from the adjacent gene zao, a master transcription factor gene pivotal for antiviral signaling. This modification process encourages the transcription factor to trigger more potent antiviral defenses in the fungal system, suggesting that these enzymes serve dual roles—modifying viral interactions while simultaneously managing the host’s response.
In experiments where the RNA interference (RNAi) pathway was disabled in N. crassa, a fascinating phenomenon emerged. The absence of this pathway led to significant growth anomalies, with increased viral transcript levels marking the infected fungi. Further exploration into the disruption of normal gene expression patterns demonstrated the upregulation of genes old-1 and old-2, both possessing deaminase domains critical for editing viral nucleotides. The deamination potentially alters the course of the infection by enhancing immune signaling pathways, thus prompting an atypical host response.
Through precise genomic analysis, it was revealed that the OLD enzymes selectively target the genomic regions upstream of old-1 and old-2. They modify the transcripts of these genes, allowing for a prolonged expression of proteins that enhance transcriptional responses. Notably, OLD-1 functions as a global RNA editor, while OLD-2 shows a more specific editing pattern on zao-2 mRNA. Such careful calibrations of RNA editing mechanisms lend itself to a broader understanding of how fungi bolster their defenses against viral infections.
The research provided intriguing insights into how altering one genetic factor can lead to unexpected viral behavior. For instance, the deletion of zao-1 in strains infected with the asymptomatic NcFV1 virus induced severe symptoms, which indicates the gene’s significant role in maintaining the asymptomatic status during viral infections. Paradoxically, further deletions altering zao-2 revitalized the fungus from symptomatic expressions, showing complex genetic interactions at play.
As the researchers investigated further, они found that in the absence of the RNAi pathway, fungi become hyperresponsive to viral infection, characterized by an overstimulated antiviral transcriptional response. Such hyper-responsiveness underscores the delicate balancing act required for survival, where appropriate genetic responses are essential for both protecting against and succumbing to viral threats.
In terms of transcriptional patterns, ZAO proteins highlighted their dual nature within the cellular context. Wild-type strains showed a less full-length expression of ZAO-1 which increased under viral pressure when RNAi was limited. These shorter variants likely curate transcriptional balance and maintain asymptomatic states by inhibiting excessive transcriptional outputs that can trigger detrimental responses, thus drawing attention to the logic underpinning fungal viral resistance.
In summary, the findings presented by Honda’s research team not only unravel the complexities of RNA editing in antifungal responses but also establish a foundational understanding that links genetic communications within fungal genomes to their viral counterparts. The evolutionary conservation of these mechanisms across diverse filamentous fungal species further endorses the significance of this research area, paving the way for future exploration to answer remaining questions.
These insights into the dynamic world of fungal virology enhance our knowledge of not only fungal biology but potentially the frameworks that underpin engineered resistance against viral pathogens. This could hold considerable implications for agricultural practices and the development of robust fungal strains with antiviral capabilities, transforming our approach to managing viral threats in crops and ecosystems alike.
By establishing a deeper understanding of how fungi navigate the complexities of viral infections through sophisticated genetic mechanisms, researchers may innovate tools to utilize these biological principles in practical applications. Thus, the ongoing studies promise to facilitate groundbreaking developments that could revolutionize the fields of plant pathology and fungal biotechnology.
In a field ripe for exploration, the research led by the University of Fukui sets a cornerstone for future inquiries into the viral world of fungi, with potential reverberations to wider scientific and agricultural contexts.
Subject of Research: Fungal antiviral responses and RNA editing mechanisms in Neurospora crassa
Article Title: RNA editing of genomic neighbors controls antiviral response in fungi
News Publication Date: 9-Apr-2025
Web References: Cell Host & Microbe
References: doi:10.1016/j.chom.2025.02.016
Image Credits: Associate Professor Shinji Honda from University of Fukui, Japan
Keywords
Tags: advancements in fungal virology researchasymptomatic viral infections in fungifungal antiviral responsesfungal virus-host dialoguegenetic engineering in mycologygenetic studies in fungal virologymechanisms of symptom induction in fungimycoviruses and host interactionsNeurospora crassa virologyRNA editing mechanisms in fungisymptom expression in viral infectionsviral infections in fungi