In a groundbreaking study poised to redefine our understanding of pancreatic cancer, researchers have unveiled a complex molecular mechanism involving HMGA2 and protein leucine methylation that orchestrates lineage plasticity within tumor cells. This breakthrough provides a compelling explanation for one of the most confounding aspects of pancreatic ductal adenocarcinoma (PDAC)—its remarkable ability to adapt and resist treatment through cellular identity shifts. As pancreatic cancer persists as one of the deadliest malignancies, insights into the biochemical drivers of its plasticity open promising avenues for targeted therapies and improved patient outcomes.
Pancreatic cancer remains notorious for its dismal prognosis, with a five-year survival rate lingering in the single digits. One major factor contributing to its lethality is the tumor’s capacity for lineage plasticity—the ability of cancer cells to switch between different cellular phenotypes, evading immune detection and developing resistance to conventional therapies. The molecular underpinnings governing this plasticity have, until now, been poorly understood. The latest research spearheaded by Dobersch, Yamamoto, Schutter, and colleagues illuminates the pivotal role played by high-mobility group AT-hook 2 (HMGA2), a chromatin-associated protein known to regulate gene expression, in steering these lineage transitions.
The scientific team meticulously demonstrated that HMGA2 operates in tandem with an epigenetic modification process known as protein leucine methylation, a relatively underappreciated post-translational modification, to drive the dynamic reprogramming of pancreatic tumor cells. Leucine methylation involves the addition of methyl groups to leucine residues on proteins, subtly altering their function and interaction landscapes. This elegant modulation adds a novel layer of regulation over cellular identity, reinforcing cancer cells’ ability to traverse diverse phenotypic states.
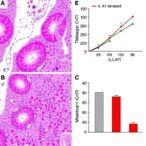
Through an array of sophisticated molecular biology techniques, including chromatin immunoprecipitation sequencing (ChIP-seq) and mass spectrometry-based proteomics, the investigators mapped the intricate interplay between HMGA2 and methylated proteins within pancreatic cancer cells. Their analyses revealed that HMGA2 recruitment to chromatin is not random but directed by the methylation status of specific transcriptional regulators and histone modifiers, which collectively reshape the epigenetic landscape to favor plasticity-associated gene expression programs.
Notably, the research uncovered that disrupting the HMGA2-protein leucine methylation axis curtails the ability of PDAC cells to switch lineages, thereby sensitizing them to standard chemotherapeutic agents such as gemcitabine. This finding has profound clinical implications, suggesting that targeting these molecular pathways could potentially subvert one of the cancer’s primary resistance mechanisms. In essence, this duo acts like a master switch, allowing cancer cells to toggle between states that enable survival under therapeutic stress.
Further mechanistic studies elucidated that HMGA2 exerts its influence by binding to AT-rich regions of DNA, which are frequently found near genes involved in epithelial-to-mesenchymal transition (EMT), stemness, and metabolic reprogramming. The protein leucine methylation modified client proteins in these pathways, leading to altered transcriptional outputs that facilitate the rewiring of the cancer cell’s identity. This suggests a highly coordinated regulatory network operates at both the chromatin structure and protein interaction levels.
Importantly, the team identified specific methyltransferase enzymes responsible for catalyzing leucine methylation on key regulatory proteins. These enzymes themselves present attractive druggable targets. The selective inhibition of these methyltransferases led to a marked reduction in tumor growth and progression in murine models, underscoring the therapeutic promise of this approach. The study’s in vivo experiments confirmed that these molecular interventions do not merely impact cell culture models but translate into tangible anti-tumor effects in living organisms.
A particularly striking aspect of this research is its potential to integrate epigenetic therapy with precision oncology. Pancreatic tumors are notorious for their genetic heterogeneity and rapid evolution. By focusing on reversible epigenetic modifications such as protein methylation and chromatin remodeling via HMGA2, therapy could be designed to dynamically adapt to tumor evolution, potentially overcoming the pervasive problem of relapse and metastasis.
The discovery also sheds light on a broader biological phenomenon—how post-translational modifications, traditionally examined in the context of phosphorylation or acetylation, can extend to methylation on non-canonical amino acids like leucine. This expands the frontier of epigenetics and proteomics research, suggesting that many other forms of subtle protein modifications may hold critical biological functions yet to be explored.
Beyond pancreatic cancer, the implications of this mechanism may ripple into other aggressive malignancies characterized by high lineage plasticity, such as small cell lung cancer and certain brain tumors. The conceptual framework provided by this study advocates for a deeper investigation into the role of HMGA2 and protein leucine methylation across diverse cancer types.
The research was facilitated by cutting-edge technology platforms, including CRISPR-based gene editing to precisely modulate HMGA2 and methyltransferase gene expression, alongside next-generation sequencing to capture the evolving transcriptomic signatures. This multi-omics approach enabled a holistic view of the molecular choreography at play, combining genomics, epigenetics, and proteomics into a coherent narrative.
Another noteworthy outcome is the potential for developing diagnostic biomarkers derived from the methylation patterns of HMGA2-associated proteins. Such biomarkers could predict a tumor’s plasticity potential and responsiveness to emerging epigenetic drugs, enabling more personalized treatment regimens and monitoring disease progression with higher accuracy.
Despite the promising advances, the authors caution that therapeutic targeting of such fundamental regulatory pathways demands precise delivery systems and a detailed understanding of off-target effects. The biological complexity and redundancy inherent in epigenetic regulation require careful clinical translation to minimize adverse effects while maximizing anti-cancer efficacy.
In summary, this pioneering investigation by Dobersch et al. elucidates a previously uncharted molecular axis—the synergistic action of HMGA2 and protein leucine methylation—as a critical driver of pancreatic cancer lineage plasticity. This paradigm-shifting insight offers new hope in tackling one of oncology’s most formidable foes by disrupting the molecular basis for tumor adaptability and therapy resistance.
As pancreatic cancer continues to defy existing treatment modalities, these findings could herald a new era of epigenetic therapy that leverages the molecular plasticity of cancer cells against them, establishing fresh strategies to improve survival rates in this devastating disease.
—
Subject of Research: Pancreatic cancer lineage plasticity driven by HMGA2 and protein leucine methylation.
Article Title: HMGA2 and protein leucine methylation drive pancreatic cancer lineage plasticity.
Article References:
Dobersch, S., Yamamoto, N., Schutter, A. et al. HMGA2 and protein leucine methylation drive pancreatic cancer lineage plasticity. Nat Commun 16, 4866 (2025). https://doi.org/10.1038/s41467-025-60129-1
Image Credits: AI Generated
Tags: biochemical drivers of tumor plasticitycellular identity shifts in cancerchromatin-associated proteins in cancerepigenetic mechanisms in PDACHMGA2 role in pancreatic cancerleucine methylation in tumorslineage plasticity in malignanciesmolecular mechanisms of cancer adaptationpancreatic cancer prognosis and survivalpancreatic ductal adenocarcinoma plasticityresistance to pancreatic cancer therapiestargeted therapies for pancreatic cancer