The link between an extra copy of chromosome 21 and Down syndrome (DS) has been well established for decades. What has not been clear was the genetic basis for the congenital heart defects that are associated with nearly half of babies born with Down syndrome. Now a new study in mice published in Nature describes how HMGN1 disrupts DNA’s packaging and regulation and how this impacts molecular levels in healthy heart development. Details of the work are published in a paper titled “Myocardial reprogramming by HMGN1 underlies heart defects in trisomy 21.”
The work is the result of a collaboration involving scientists from Gladstone Institutes, Sanford Burnham Prebys, and elsewhere. As explained in the paper, the link to HMGN1 was made using human pluripotent stem cell and mouse models of Down syndrome. Specifically, “single-cell transcriptomics showed that trisomy 21 shifts human [atrioventricular canal] cardiomyocytes towards a ventricular cardiomyocyte state,” the scientists wrote. Then, “a CRISPR-activation single-cell RNA droplet sequencing screen of chromosome 21 genes expressed during heart development revealed that HMGN1 upregulation mimics this shift, whereas deletion on one HMGN1 allele in trisomic cells restored normal gene expression.”
According to Sanjeev Ranade, PhD, assistant professor in the Center for Cardiovascular and Muscular Diseases and Center for Data Science and Artificial Intelligence at Sanford Burnham Prebys,“what our paper did was address a major unresolved question: Yes, three copies of chromosome 21 causes DS, but why? What are the genes on chromosome 21 that are bad if you have them in three copies? How in the world do you try to find those genes?” Ranade is the first author on the paper and also a co-corresponding author.
While this study was done in mice, there are obvious benefits for research in people. Learnings from this study “could pave the way for treatments to help prevent heart malformations in people with Down syndrome and related heart defects, which would be a major win for patients and their families,” said Deepak Srivastava, MD, president and senior investigator at Gladstone, a pediatric cardiologist at University of California, San Francisco (UCSF). Srivastava is the senior author on the paper and one of its corresponding authors.
Digging into the details, Srivastava, Ranade, and their colleagues focused on individuals who are mosaic for trisomy 21, a rare situation where some cells in the body have three copies of chromosome 21 while other cells have two. Mosaic individuals do not have features of Down syndrome but are at high risk of having children with the condition.
Working with this population made sense because it eliminated a potential source of uncertainty. In most cases, researchers comparing healthy and Down syndrome cells have to collect cells from two individuals. This is problematic because when they observe differences in how cells function, it is unclear whether these are due to chromosome 21 or to other types of genetic variations. Working with cells from mosaic people clears up that question. “Everything else in these cells was exactly the same, so this gave us a perfect, natural control experiment,” Ranade said.
The scientists took cell samples from mosaic individuals and used induced pluripotent stem cell technology to transform them into heart cells. “The aha moment came when we found a striking difference in the heart cells with three copies of chromosome 21, compared to those with two copies,” Srivastava said. “That made us wonder which gene on the chromosome was causing this dramatic shift in the cells.”
That’s when the researchers turned to CRISPR to boost the levels of candidate genes on chromosome 21. Working with one at a time, they activated the genes in healthy cells containing the normal two copies of the chromosome to see if any mimicked the problems seen in trisomy cells. They then used artificial intelligence to model the differences between healthy heart cells and those impacted by Down syndrome.
“The model allowed us to gain insights from the huge amount of data generated by the lab experiments,” explained Sean Whalen, PhD, a co-author on the study and principal staff research scientist at Gladstone Institutes. “As a result, we were able to predict that one gene, called HMGN1, made heart cells look like the abnormal cells in Down syndrome. Surprisingly, this gene wasn’t on anyone’s radar as one that could cause the disorder.”
Once they identified the relevant gene in cells, the scientists moved on to validating their finding in a mouse model of Down syndrome. In these animals, “when we reduced the levels of HMGN1, heart defects disappeared,” according to Feiya Li, PhD, a postdoctoral fellow in Srivastava’s lab and co-first author of the study. “It was a powerful indication that three copies of this gene are needed to cause the heart defects in Down syndrome.”
While increases in HMGN1 expression may be necessary for cardiac defects in Down syndrome cases, there is evidence of involvement from other genes including one dubbed DYRK1. As part of their next steps, Srivastava’s lab is testing whether combining the two is sufficient to cause cardiac defects in mice.
Overall, “our work shows the power of combining cutting-edge genomics with advanced computational modeling,” said Katie Pollard, PhD, director of the Gladstone Institute of Data Science and Biotechnology, professor at UCSF, and co-author on the study. “We now have a roadmap not only for understanding heart defects in Down syndrome, but for studying the root causes of other diseases caused by extra or missing chromosomes.”
Ranade added, “we’re hoping that our approach in this paper lays out a roadmap for finding genes driving other kinds of defects, such as intellectual disability or in bone formation, seen in children with Down syndrome. Ultimately, once we find the players that drive disease, we believe we can use that information to find drugs that could help people with Down syndrome.”

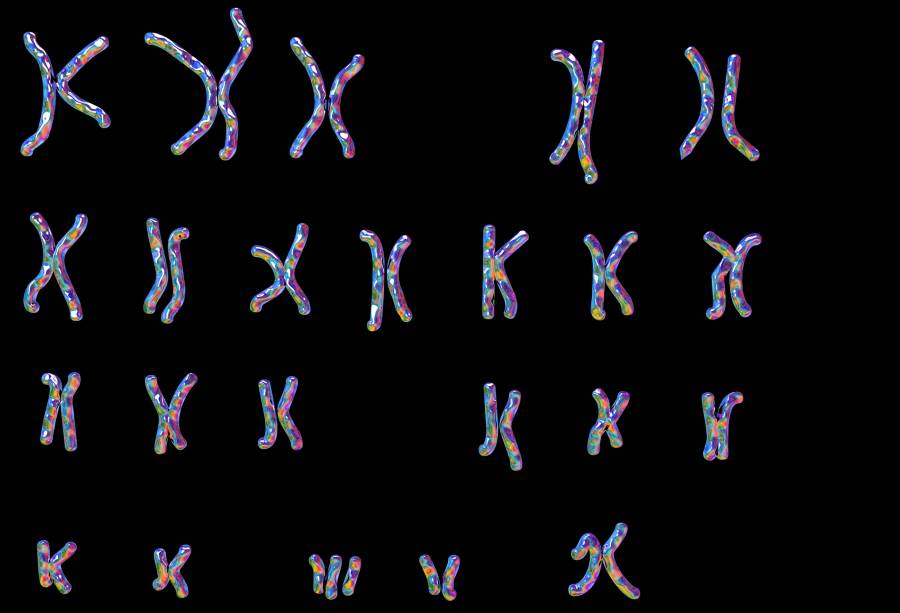
![GettyImages-685025133_edited Karyotype showing the arrangement of chromosomes in a female with Downs syndrome (trisomy-21), the most common diagnosable cause of mental handicap, computer illustration. Downs syndrome is caused by a chromosomal anomaly: the 21st set having three rather than the normal two chromosomes. In karyotyping, chromosomes are arranged in numbered pairs according to a standard classification. The female set differs from the male in the sex pair (bottom right); a male would have XY rather than XX. A female Downs karyotype is written: 47, XX, +21. The normal chromosome count is 46, 23 from maternal and 23 from paternal origin [ KATERYNA KON/SCIENCE PHOTO LIBRARY]](https://www.genengnews.com/wp-content/uploads/2025/10/GettyImages-685025133_edited-696x474.jpg)