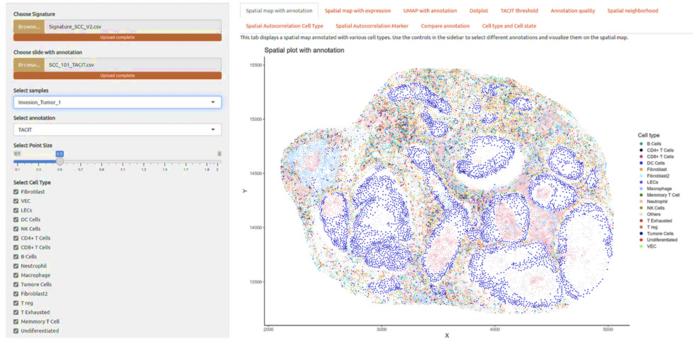
In a groundbreaking advancement at the intersection of artificial intelligence and biomedical research, scientists at Virginia Commonwealth University’s Massey Comprehensive Cancer Center have developed an innovative algorithm named TACIT (Threshold-based Assignment of Cell Types from Multiplexed Imaging Data). This novel computational tool dramatically accelerates the identification and classification of cells within complex tissues, offering a quantum leap in the speed and precision with which researchers and clinicians can analyze cellular environments. Published recently in Nature Communications, this breakthrough holds tremendous promise not only for cancer treatment but for broad applications across medicine and pharmacology.
TACIT was designed to address a critical bottleneck in cell biology and clinical diagnostics: the labor-intensive and time-consuming process of cell type assignment from multiplexed imaging data. Traditional methods often rely on a limited set of biomarkers to distinguish cell types and states, frequently resulting in ambiguous or incomplete analyses. By leveraging advanced machine learning techniques and artificial intelligence, TACIT can parse the expression profiles of millions of cells across multiple tissues—such as brain, gut, and oral glands—with unprecedented accuracy and speed, reducing the typical analysis time from over a month to mere minutes.
The core of TACIT’s capability lies in its use of marker-expression thresholds that allow for the precise annotation of cells based on their protein and RNA signatures. Unlike conventional unsupervised clustering techniques, TACIT incorporates spatial multiomics, integrating both transcriptomic and proteomic data in situ. This integration empowers the algorithm to decipher subtle variations in cell states and interactions, rendering a detailed and nuanced cellular map that was previously unattainable at scale. Such rich data synthesis paves the way for enhanced biomarker discovery and a deeper understanding of tissue biology.
.adsslot_boSgv3EcNn{width:728px !important;height:90px !important;}
@media(max-width:1199px){ .adsslot_boSgv3EcNn{width:468px !important;height:60px !important;}
}
@media(max-width:767px){ .adsslot_boSgv3EcNn{width:320px !important;height:50px !important;}
}
ADVERTISEMENT
Developed through the collaboration between Dr. Jinze Liu, a professor of Biostatistics at VCU’s School of Public Health, and Dr. Kevin Byrd, an assistant professor at the School of Dentistry, TACIT embodies a fusion of computational rigor and biological insight. The duo utilized data derived from over five million cells, creating a robust and highly extensible framework. This vast dataset, encompassing diverse organ systems and cell populations, provides TACIT the capacity to generalize effectively beyond any one tissue or disease context, effectively offering a universal key to decoding cellular heterogeneity.
The implications of TACIT for cancer diagnosis and treatment are nothing short of transformative. By enabling rapid and highly accurate cell identification, clinicians can more quickly pinpoint malignant versus healthy cell populations and better characterize the tumor microenvironment. This accelerated diagnostic precision supports tailored therapeutic decisions, ensuring patients receive the most effective treatments sooner and potentially sparing them from ineffective or unnecessary interventions. Furthermore, TACIT’s spatial biology prowess can illuminate new cellular pathways and interactions that underlie cancer progression and resistance.
On a technical level, TACIT outperforms existing unsupervised cell annotation methods by harmonizing proteomic and genetic data streams to enhance reliability. Its algorithmic design ensures scalability, capable of handling increasing amounts of data without compromising speed or accuracy. This quality is crucial as spatial multiomic technologies proliferate, generating ever-larger datasets. The adaptability of TACIT to grow with data availability means it can continually refine its predictive power and diagnostic utility, benefiting from iterative learning.
Beyond cancer, TACIT’s versatility extends into pharmacological research and clinical trial optimization. A major obstacle in trials is the heterogeneous patient response to experimental therapies, often due to insufficient biomarkers that predict efficacy. TACIT’s ability to identify nuanced spatial biomarkers allows for preemptive stratification of trial participants, matching the right candidates to the right interventions. This precision not only enhances trial success rates but also spares ineligible patients from ineffective regimens, representing a paradigm shift in personalized medicine.
The algorithm also incorporates RNA marker data, enabling insights into gene expression patterns that correlate with drug responsiveness. By mapping these molecular profiles to a comprehensive repository of FDA-approved pharmaceuticals, TACIT offers the tantalizing prospect of repurposing existing drugs based on a patient’s unique tissue microenvironment. This drug mapping feature could significantly streamline treatment decisions, providing more therapeutic options when conventional paths falter and reducing the need for new investigational drugs when existing ones suffice.
TACIT’s multi-modal approach is another key innovation. The researchers have demonstrated a new technique linking slide proteomics with transfer proteomics, effectively producing cell multi-omics datasets where multiple markers are studied simultaneously at the single-cell level. Prior to this development, researchers were mostly constrained to single-omics approaches, limiting the depth of insights attainable. This multi-omics integration unlocks a richer biological context, revealing interactions across different biomolecular layers that govern cell behavior and disease processes.
Liu and Byrd liken TACIT to a “Rosetta Stone” for spatial biology, translating disparate data types into a unified language that accelerates discovery and clinical translation. By bridging protein, RNA, and spatial information, TACIT enables researchers to unlock complex biological codes and discern cell relationships that were previously hidden. This capacity holds promise not only for oncology but also for neurobiology, immunology, and other areas where cellular diversity and organization critically influence health and disease.
The future trajectory for TACIT envisions continuous expansion and refinement. As more datasets are incorporated, and as spatial multiomics technologies evolve, the algorithm will become even more powerful. Integration with emerging imaging platforms and artificial intelligence tools will further enhance its diagnostic accuracy and ease of use. Coupling TACIT with clinical workflows promises to revolutionize precision medicine, providing actionable insights that advance patient care.
This breakthrough underscores the critical role of interdisciplinary collaboration in modern biomedical innovation—merging statistics, computer science, molecular biology, and clinical expertise to solve fundamental challenges. TACIT’s rapid deployment could redefine standards for spatial biology and accelerate the translation of complex tissue data into meaningful, patient-centered outcomes. It heralds an era where computational algorithms not only augment human expertise but become indispensable partners in the quest to understand and treat disease.
Virginia Commonwealth University’s commitment to cutting-edge research is exemplified in the development of TACIT, which has garnered support from prestigious funders including the Chan Zuckerberg Initiative, ADA Foundation, and the National Cancer Institute. The work is poised to inspire further advances across the biomedical sciences, establishing a new gold standard for cellular characterization and genomic medicine.
In summary, TACIT is more than an algorithm; it represents a paradigm shift in how we visualize, interpret, and intervene in complex biological systems. Its ability to speed up cell type identification by orders of magnitude promises to transform diagnostics, therapeutic stratification, and drug discovery. As spatial multiomics continues to generate rich, multi-layered data, tools like TACIT will be crucial for unlocking their full potential and ultimately improving patient outcomes worldwide.
Subject of Research:
Artificial Intelligence-Driven Cellular Deconvolution and Spatial Multiomics Analysis for Biomedical Applications
Article Title:
Deconvolution of cell types and states in spatial multiomics utilizing TACIT
News Publication Date:
21-Apr-2025
Web References:
https://www.nature.com/articles/s41467-025-58874-4
References:
Liu, J., Byrd, K. et al. Deconvolution of cell types and states in spatial multiomics utilizing TACIT. Nature Communications (2025). DOI: 10.1038/s41467-025-58874-4
Image Credits:
Virginia Commonwealth University
Keywords:
Artificial intelligence, Cancer, Live cell imaging, Imaging, Medical imaging, Genetic algorithms, Biomarkers, Cell biology
Tags: advanced cell classification techniquesartificial intelligence in biomedical researchbreakthroughs in clinical diagnosticscomputational tools for cell biologyenhancing pharmacological research methodsimproving cancer treatment methodologiesmachine learning for cancer treatmentmultiplexed imaging data analysisspeeding up cellular environment analysisTACIT algorithm in cancer diagnosistransformative technology in medicineVCU Massey Comprehensive Cancer Center innovations