Breast cancer stands as one of the foremost health challenges impacting women globally, and despite significant advances in diagnosis and treatment, certain aggressive subtypes continue to evade effective therapeutic intervention. Among these, triple-negative breast cancer (TNBC) remains particularly formidable. Representing approximately 10 to 15 percent of breast cancer diagnoses, TNBC disproportionately affects younger women and African American populations, with a clinical profile marked by rapid progression and limited treatment options. Unlike hormone receptor-positive breast cancers, TNBC lacks expression of estrogen receptor, progesterone receptor, and HER2, rendering conventional targeted therapies ineffective. This alarming gap underscores an urgent need for novel molecular insights that could pave the way for innovative therapeutic strategies against this aggressive malignancy.
A groundbreaking study emerging from the Cold Spring Harbor Laboratory (CSHL) reveals a promising breakthrough in understanding the molecular underpinnings of TNBC. Led by Professor David Spector and graduate researcher Wenbo Xu, the team has unveiled the critical role of a long non-coding RNA (lncRNA) known as LINC01235 in the regulation of TNBC tumorigenesis. Published in the journal Molecular Cancer Research, their research delineates how LINC01235 serves as an upstream regulator of the NFIB gene and the NOTCH signaling pathway, both pivotal components implicated in cancer cell proliferation and progression. This discovery signals a significant advancement in the quest to identify molecular targets for TNBC, potentially marking a new chapter in breast cancer therapeutics.
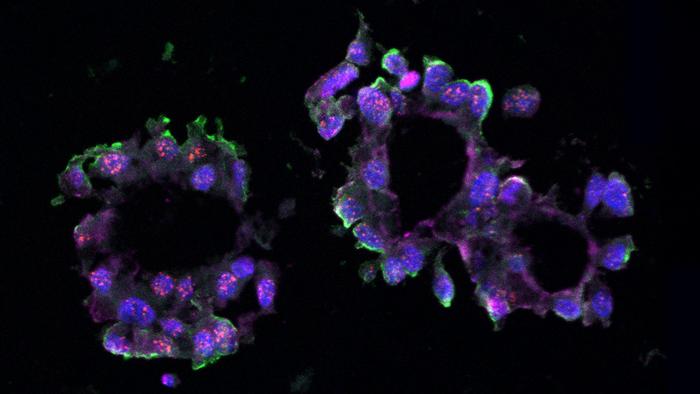
Long non-coding RNAs, once dismissed as “junk” DNA components, have rapidly ascended into the spotlight of molecular oncology due to their diverse regulatory roles in gene expression, chromatin remodeling, and cellular signaling. The Spector laboratory has long specialized in decoding the functions of lncRNAs in cancer biology. Their recent focus on LINC01235 emerged from comprehensive RNA sequencing analyses performed on TNBC organoids — miniature, three-dimensional cell culture models mimicking the structural and functional complexity of human tumors. These organoids, derived from patient donated tissue samples, provide a robust platform to investigate tumor-specific molecular interactions in an environment closely resembling in vivo conditions.
.adsslot_fAClVuPnHy{ width:728px !important; height:90px !important; }
@media (max-width:1199px) { .adsslot_fAClVuPnHy{ width:468px !important; height:60px !important; } }
@media (max-width:767px) { .adsslot_fAClVuPnHy{ width:320px !important; height:50px !important; } }
ADVERTISEMENT
The investigative team’s hypothesis took shape upon mining data from The Cancer Genome Atlas, encompassing genomic profiles of over 11,000 cancer patients. Bioinformatic correlation revealed a significant positive relationship between LINC01235 and NFIB, a nuclear factor previously implicated in breast cancer aggressiveness yet not fully characterized in TNBC. This observation propelled extensive functional studies aimed at dissecting the mechanistic role of LINC01235 within TNBC cellular frameworks. Until now, both LINC01235 and NFIB had remained relatively uncharted in the context of triple-negative pathology, making this foray a pioneering step in uncovering novel regulatory networks driving oncogenesis.
To probe the functional relevance of LINC01235, the researchers employed CRISPR-Cas9 gene editing technology to knockout its expression in cultured TNBC cells. Parallel experiments utilized antisense oligonucleotides to knockdown LINC01235 levels both in traditional cell cultures and in three-dimensional organoids. Remarkably, both intervention strategies resulted in a significant downregulation of NFIB expression, accompanied by a marked suppression of organoid formation and cellular proliferation. This evidence strongly supports a model wherein LINC01235 exerts a direct positive regulatory influence on NFIB transcription, thereby sustaining the malignant phenotype characteristic of TNBC.
Delving deeper into the pathway-level consequences of this RNA-mediated regulation, the team identified the NOTCH signaling cascade as a critical downstream effector impacted by LINC01235 and NFIB activity. The NOTCH pathway, well-established in governing cell fate decisions, differentiation, and proliferation, has been extensively linked to cancer stemness and chemoresistance. Xu explains that the modulation of NOTCH signaling by LINC01235-NFIB axis influences TNBC cell proliferation, effectively contributing to tumor growth and maintenance. Given that aberrant NOTCH signaling is a hallmark in various malignancies, the identification of upstream lncRNA regulators presents a novel dimension for therapeutic targeting.
This discovery carries significant clinical implications. The current therapeutic landscape for TNBC remains largely restricted to chemotherapy and radiotherapy, modalities burdened by toxicities and oftentimes suboptimal efficacy due to intrinsic tumor heterogeneity. Targeting non-coding RNAs such as LINC01235 represents an innovative strategy, potentially enabling precision medicine approaches that disrupt fundamental molecular circuits sustaining the cancer. Additionally, antisense technologies or RNA-based therapeutics could be exploited to selectively silence oncogenic lncRNAs, opening exciting avenues for translational research and drug development in aggressive breast cancers.
However, the authors emphasize that much work remains before clinical applications can be realized. Comprehensive validation in preclinical models, detailed mapping of interaction partners, and elucidation of downstream regulatory networks are essential to fully harness the therapeutic potential of LINC01235. Furthermore, understanding the dynamics of lncRNA expression across diverse TNBC patient populations could shed light on predictive biomarkers for treatment response and disease prognosis. This foundational research thus sets the stage for expansive multidisciplinary efforts combining molecular biology, bioinformatics, and clinical oncology to translate these insights into tangible benefits for patients.
The utilization of patient-derived organoids as a model system underscores the evolving landscape of cancer research methodologies, bridging the gap between simplistic cell line studies and complex in vivo investigations. These organoids preserve the genetic heterogeneity and microenvironmental interactions of original tumors, enabling high-fidelity studies of tumor biology and drug responsiveness. The success of the Spector lab in deploying this platform to uncover lncRNA-mediated regulatory mechanisms exemplifies the powerful synergy of advanced genomic tools and physiologically relevant experimental systems in driving cancer discoveries.
Moreover, this study exemplifies the expanding recognition of the non-coding genome’s role in health and disease. While protein-coding genes account for a fraction of the human genome, non-coding elements, particularly lncRNAs, represent a vast and largely untapped reservoir of regulatory complexity. The nuanced regulatory roles of lncRNAs, including epigenetic modulation, RNA-protein interactions, and scaffolding functions, position them as critical nodes controlling cellular phenotypes. Evidence increasingly implicates their dysregulation in oncogenesis, metastasis, and therapy resistance, making them an exciting frontier in cancer biology and therapeutics.
As Professor Spector poignantly notes, the ultimate goal is to unravel the molecular mechanisms whereby cells maintain normal functions and how disease states usurp these controls, oftentimes via dysregulated RNA molecules. LINC01235 serves as a representative piece in this intricate puzzle—highlighting how subtle shifts in RNA expression profiles can drive profound pathological changes. Long-term, targeting such lncRNAs offers hope for therapies that are not only effective but also possess a degree of specificity that minimizes detrimental off-target effects typical of conventional anti-cancer drugs.
In closing, the identification of LINC01235 as an upstream regulator of NFIB and the NOTCH pathway in TNBC represents a pivotal advance in breast cancer research. It underscores the importance of exploring RNA-mediated gene regulatory networks and leveraging contemporary technologies like CRISPR and organoid culture systems to reveal novel vulnerabilities in notoriously difficult cancers. This research ignites optimism that innovative molecular targets are within reach, propelling the field closer to the development of effective interventions against triple-negative breast cancer.
Subject of Research: Role of long non-coding RNA LINC01235 in triple-negative breast cancer through regulation of NFIB and NOTCH pathway
Article Title: LINC01235 is an Upstream Regulator of the NFIB Gene and the NOTCH Pathway in Triple Negative Breast Cancer
News Publication Date: 30-Jun-2025
Web References: http://dx.doi.org/10.1158/1541-7786.MCR-24-1143
References: Published in Molecular Cancer Research, American Association for Cancer Research
Image Credits: Spector lab / Cold Spring Harbor Laboratory
Keywords: Long noncoding RNA, Breast cancer, Progenitor cells, Notch pathway, Organoids
Tags: African American breast cancer statisticsaggressive breast cancer subtypescancer treatment breakthroughsCold Spring Harbor Laboratory researchdisparities in breast cancer incidenceinnovative therapies for TNBClong non-coding RNA in TNBCmolecular insights in cancer treatmentNFIB gene regulation in breast cancerNOTCH signaling pathway in oncologytriple-negative breast cancer researchyoung women and breast cancer