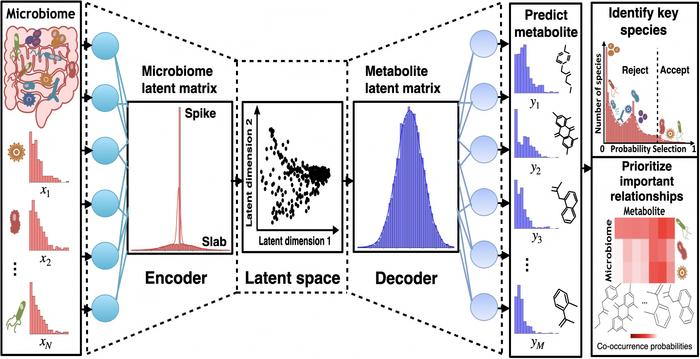
In a groundbreaking leap for microbiome research and computational biology, scientists at the University of Tokyo have developed an advanced artificial intelligence framework named VBayesMM, poised to revolutionize our understanding of the complex interactions between gut bacteria and human metabolites. By leveraging a state-of-the-art variational Bayesian neural network, the team has tackled one of the most daunting challenges in biomedical science: decoding the high-dimensional and multifaceted relationships within microbiome multiomics data. This novel approach transcends the limitations of traditional analytical models by effectively prioritizing crucial microbial-metabolite interactions, thereby opening new frontiers for personalized medicine and targeted therapies.
The human gut is home to a staggering population of bacteria—approximately 100 trillion microorganisms, far outnumbering the human cells within our own bodies. These bacteria are not mere passengers but active participants influencing a wide array of physiological processes. They are involved not only in digestion but also modulate immune responses, metabolic pathways, neural functions, and even mood states. Central to these effects are metabolites, small molecules produced or modified by gut microbes that serve as molecular messengers throughout the body. Understanding which specific bacterial species influence particular metabolites—and how these relationships shift in various health conditions—has been a scientific holy grail. Yet, the sheer diversity and complexity of microbial populations and their chemical products have rendered traditional analytical strategies inadequate.
Enter VBayesMM, an AI-powered system designed to navigate the labyrinth of microbiome-driven metabolic interactions. Unlike conventional methods that may overfit data or produce overconfident predictions, this Bayesian neural network inherently models uncertainty, allowing researchers to discern not only the presence but also the reliability of inferred relationships. By integrating paired microbiome and metabolite datasets, VBayesMM distinguishes key microbial contributors amidst a vast sea of background noise. This discriminative capacity is critical when handling thousands of microbial species alongside an equally vast array of metabolites, scenarios where common machine learning frameworks often falter.
.adsslot_13hLOQbDXF{width:728px !important;height:90px !important;}
@media(max-width:1199px){ .adsslot_13hLOQbDXF{width:468px !important;height:60px !important;}
}
@media(max-width:767px){ .adsslot_13hLOQbDXF{width:320px !important;height:50px !important;}
}
ADVERTISEMENT
Project researcher Tung Dang and his colleagues meticulously tested VBayesMM against real-world datasets encompassing diverse health conditions such as sleep disorders, obesity, and cancer. In each case, the model surpassed existing computational methods in accuracy and biological relevance. Notably, the bacteria identified by VBayesMM aligned with known physiological roles, bolstering confidence in the approach’s validity. This robust performance underscores the system’s potential not simply as a data analysis tool but as a discovery engine capable of uncovering hitherto hidden microbial-metabolite dynamics integral to disease pathology and wellness.
One of the critical advancements VBayesMM brings is its ability to quantify uncertainty in predictions, an often-overlooked aspect in AI-driven biomedical analyses. By modeling uncertainty, the network prevents overinterpretation of tentative associations that might lead researchers astray. This fosters a more cautious and scientifically rigorous interpretation of high-dimensional data—where spurious correlations are abundant—and thus paves the way for more reliable biomarker identification and therapeutic targeting.
Despite its promising capabilities, VBayesMM is not without limitations. The approach currently benefits from datasets richer in microbial abundance profiles than in metabolite measurements, leading to reduced predictive accuracy when bacterial data is sparse. Additionally, the model assumes independence among microbial species, whereas in reality, gut bacteria engage in complex, synergistic, and competitive interactions forming an intricate ecological network. Capturing such interdependencies constitutes an ongoing challenge poised to be addressed through future refinements integrating microbial phylogenetic relationships and interaction networks.
Looking ahead, the research team aspires to incorporate more comprehensive chemical datasets encompassing a broader spectrum of bacterial products. This expansion faces significant hurdles in distinguishing metabolites derived from microbes versus those originating from host metabolism or external dietary inputs. Enhancing the robustness of VBayesMM across diverse patient populations is another priority, acknowledging the vast variability in microbiomes shaped by genetics, environment, lifestyle, and geography.
Moreover, computational efficiency remains an important consideration. Mining multiomic datasets—encompassing tens of thousands of microbial species and metabolites—demands substantial processing power and time. Although VBayesMM is optimized for high-throughput workloads, continued algorithmic improvements and hardware advancements will be essential to facilitate widespread adoption by the scientific community and clinical practitioners.
The long-term vision driving this endeavor is profoundly translational: enabling precision medicine interventions that manipulate gut microbiota to yield therapeutic benefits. Whether through cultivating select bacterial strains that produce beneficial metabolites or designing drugs that modulate metabolite levels, these insights could transform the management of complex diseases such as metabolic disorders, neuropsychiatric conditions, and cancer. By elucidating the hidden molecular dialogues between our microbial partners and ourselves, VBayesMM charts a course toward harnessing the microbiome as an actionable component of health care.
Tung Dang eloquently emphasized the significance of their work: “By accurately mapping these bacteria-chemical relationships, we could potentially develop personalized treatments. Imagine being able to grow a specific bacterium to produce beneficial human metabolites or designing targeted therapies that modify these metabolites to treat diseases.” This vision reflects a paradigm shift where AI-driven personalized microbiome analytics integrate seamlessly into clinical decision-making, ushering in an era of bespoke therapeutics.
This research exemplifies the power of interdisciplinary collaboration, combining expertise in computational modeling, microbiology, metabolomics, and clinical science. Supported by prominent funding agencies including the Japan Society for the Promotion of Science (JSPS) and JST CREST, the project stands at the intersection of cutting-edge AI and biological inquiry. It highlights the escalating role of computational simulations and machine learning techniques in resolving biological complexity, a trend that will undoubtedly accelerate biomedical discovery in years to come.
Ultimately, VBayesMM not only innovates methodologically but also addresses a core challenge in multiomics data analysis: making sense of high-dimensional datasets rife with noise and uncertainty. Its Bayesian neural network framework may serve as a template for analogous problems in other biological domains, such as cancer genomics or immunology, where the interplay of myriad variables demands sophisticated probabilistic modeling.
As large-scale microbiome projects continue to generate exponentially increasing datasets, tools like VBayesMM will be indispensable for unlocking their full potential. While technical refinements and expanded datasets await, this pioneering approach sets a new benchmark for microbiome analytics and illustrates how artificial intelligence can deepen our understanding of the microscopic worlds within us.
Subject of Research: Cells
Article Title: VBayesMM: Variational Bayesian neural network to prioritize important relationships of high-dimensional microbiome multiomics data
News Publication Date: 4-Jul-2025
References: Tung Dang, Artem Lysenko, Keith A. Boroevich and Tatsuhiko Tsunoda, “VBayesMM: Variational Bayesian neural network to prioritize important relationships of high-dimensional microbiome multiomics data”, Briefings in Bioinformatics, DOI: 10.1093/bib/bbaf300
Image Credits: ©2025 Tsunoda et al. CC-BY-ND
Keywords
Bayesian neural network, microbiome, metabolites, gut bacteria, multiomics, artificial intelligence, computational biology, metabolomics, microbiome-host interactions, personalized medicine, probabilistic modeling, microbial ecology
Tags: artificial intelligence in microbiome researchcomputational biology innovationsdecoding complex biological relationshipsgut bacteria and human metabolitesgut health and disease connectionsinteractions between gut microbiota and metabolitesmicrobial metabolism and human healthmicrobiome multiomics data analysispersonalized medicine advancementstargeted therapies in gut healthUniversity of Tokyo microbiome studyvariational Bayesian neural network applications